
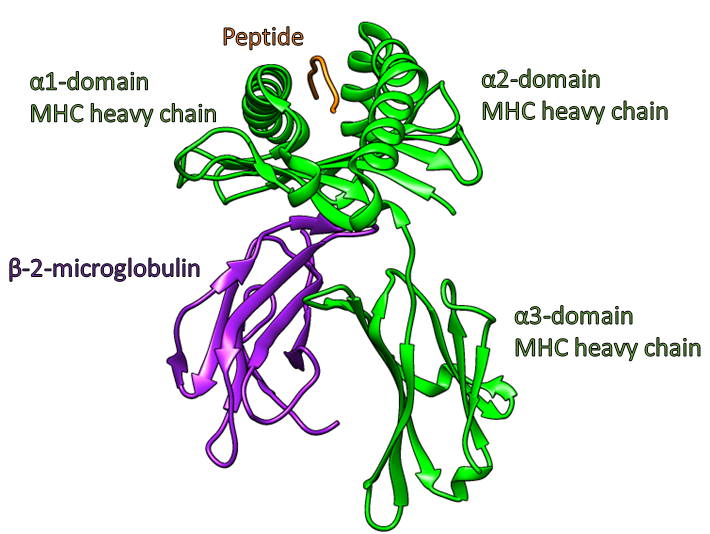
I am currently working to investigate the conformational dynamics of MHC-I proteins by combining experimental hydrogen-deuterium exchange (HDX) datasets and computational molecular dynamics simulations. HDX is limited as deuterium exchange is measured across entire peptide fragments leading to low resolution qualitative analysis, preventing insight at the single residue level. By combining the atomistic resolution conformational ensembles sampled by MHC proteins during molecular dynamics simulations we can predict the probable structures present in the HDX experiments.To this end, I will be using a variety of enhanced sampling methods and experimentally directed simulation biasing techniques to explore the mechanistic functions of MHC-I, both independently and within the broader context of the peptide loading complex. This will help provide insight into how different MHC alleles are able to select for certain peptides ligands and improve our ability to predict tumour specific antigens for use as personalised cancer vaccine targets.