
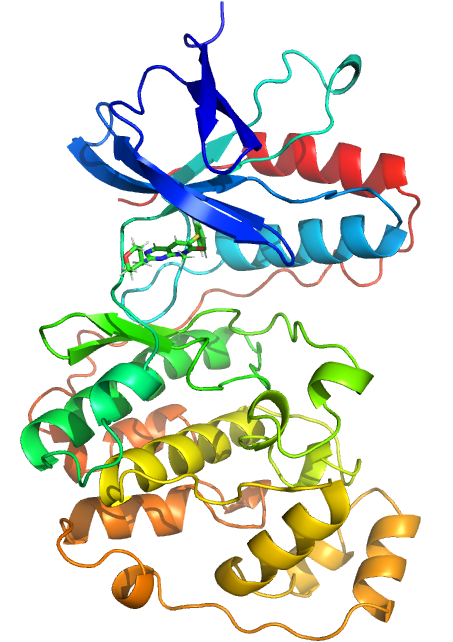
My project is centred around calculating relative binding free energies using alchemical perturbation methods with focus on application to drug discovery. Currently, I am working on improving the free energy estimations for a congeneric series of drug-like inhibitors binding to P38 Mitogen Activated Protein Kinase, which is a target of interest in the pharmaceutical industry as it influences the inflammatory immune response of the body.This is of particular relevance when considering diseases such as Rheumatoid Arthritis and Crohn’s Disease. Free energy methods are powerful tools and their use is becoming ever more popular in the drug disovery context, yet a significant drawback of their usage is the ability of the computational estimates to reliably and accurately emulate experimental data. To address this, I will be investigating how the forcefields used in the simulations are influencing the sampling of the ligand binding dynamics and using these insights to modify existing forcefield parameters, with the intention of obtaining free energy estimates which are reliably within 1 kcal mol-1 of experiment.